Research
Role of non-coding RNA in gene expression control
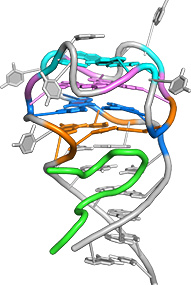
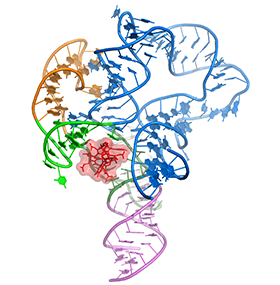
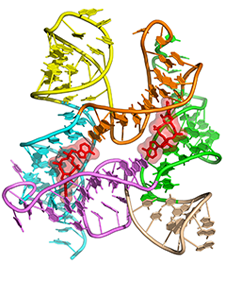
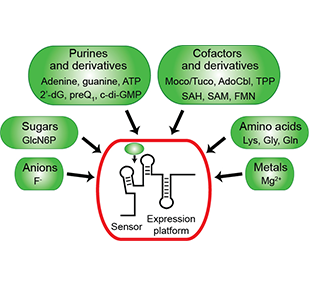
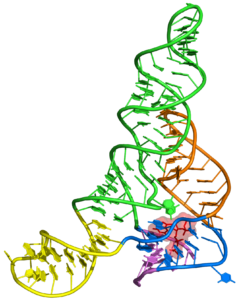
One of the main goals of our laboratory is to understand the mechanistic principles underlying gene expression control by non-coding RNAs in both prokaryotes and eukaryotes. In our bacterial studies, we focus on a novel class of RNA regulators termed riboswitches. Riboswitches primarily control the expression of numerous metabolic pathways in response to elevated concentrations of specific cellular metabolites. Riboswitches adopt well-defined spatial structures tuned for selective recognition of cognate small molecule ligands. To understand the molecular basis of gene expression control by riboswitches, We have determined crystal structures of abundant riboswitches that sense adenosylcobalamin, the active form of vitamin B12, c-di-AMP, a cyclic dinucleotide second messenger, PRPP, an intermediate in the purine biosynthesis pathway, and ppGpp, a famous bacterial alarmone. We are interested in how riboswitches discriminate cognate ligands from other cellular metabolites and how they regulate gene expression. Some riboswitches control genetic circuits essential for viability of bacteria and are therefore ideally suited for binding to analogs of natural ligands which could shut down associated pathways and kill pathogenic bacteria.
Our eukaryotic studies are centered on the molecular mechanisms of transcriptional control mediated by long non-coding RNAs (lncRNA). We are particularly interested in how lncRNA contributes to the activation of inflammatory genes and why mutations in lncRNA cause human diseases.
 |
 |
 |
 |
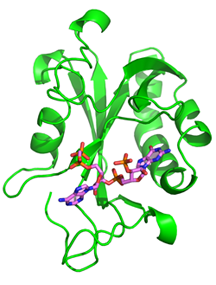
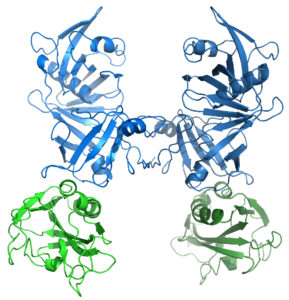
One such devastating neurological disease, Fragile X syndrome (FXS), results from the loss of the RNA-binding protein Fragile Mental Retardation Protein (FMRP). FXS is the most common single-gene disorder, the most frequent cause of inherited intellectual disability, and the most common monogenetic cause of autism. FMRP interacts with a subset of neuronal mRNAs and controls their translation in synapses. This control contributes to changes in neuronal circuits and affects learning and memory. We pursue structural and biochemical studies to understand the mechanisms of mRNA selection and translational control by FMRP and determine the molecular basis of FXS and related disorders.
Mechanisms of mRNA modification and degradation
Although many regulatory circuits involve transcriptional control, posttranscriptional events such as RNA degradation could have a comparably strong impact on the well-being of organisms by directly modulating protein biosynthesis through availability of mRNAs for translation. We are particularly interested in understanding the early events that trigger mRNA degradation in bacteria. We are also elucidating the molecular mechanisms of mRNA modification and impact of modifications on mRNA degradation and other cellular activities in bacteria and animals.
Mechanism of transcription in bacteria
RNA transcription is a keystone cellular process in all organisms. Transcription proceeds through several steps and requires concerted action of RNA polymerase and many other factors. We envision that determining three-dimensional structures of complexes between RNA polymerase and ancillary proteins will be essential to understand molecular mechanisms of transcription and identify novel antibacterial targets. Our current efforts are focused on determining the spatial structure of macromolecular assemblies associated with transcription in bacteria by cryo-EM.